Graph Module
The synkit.Graph package provides core graph-based utilities in three submodules:
Matcher: graph comparison and subgraph search
ITS: Internal Transition State graph construction and decomposition
MTG: Mechanistic Transition Graph generation and exploration
Graph Canonicalization
The class GraphCanonicaliser canonicalises a graph by computing a canonical relabeling of node indices. It employs a Weisfeiler–Lehman colour-refinement backend (default: 3 iterations) to ensure that each atom-map assignment is uniquely and consistently ordered across isomorphic reactions [1].
1from synkit.IO import rsmi_to_its
2from synkit.Graph.canon_graph import GraphCanonicaliser
3from synkit.Graph.Matcher.graph_matcher import GraphMatcherEngine
4
5canon = GraphCanonicaliser(backend='wl', wl_iterations=3)
6rsmi = (
7 '[CH3:1][CH:2]=[O:3].'
8 '[CH:4]([H:7])([H:8])[CH:5]=[O:6]>>'
9 '[CH3:1][CH:2]=[CH:4][CH:5]=[O:6].'
10 '[O:3]([H:7])([H:8])'
11)
12its_graph = rsmi_to_its(rsmi)
13canon_graph = canon.canonicalise_graph(its_graph).canonical_graph
14print(its_graph == canon_graph) # False
15
16gm = GraphMatcherEngine(backend='nx')
17print(gm.isomorphic(its_graph, canon_graph)) # True
Matcher
The synkit.Graph.Matcher submodule offers:
GraphMatcherEngine— generic graph-isomorphism and subgraph matchingSubgraphMatch— subgraph search
Example: Graph Isomorphism
Check whether two ITS graphs—derived from reaction SMILES differing only by atom‐map ordering—are truly isomorphic:
1from synkit.IO import rsmi_to_its
2from synkit.Graph.Matcher.graph_matcher import GraphMatcherEngine
3
4# Two reaction SMILES with permuted atom-map labels
5rsmi_1 = (
6 '[CH3:1][C:2](=[O:3])[OH:4].[CH3:5][OH:6]'
7 '>>'
8 '[CH3:1][C:2](=[O:3])[O:6][CH3:5].[OH2:4]'
9)
10rsmi_2 = (
11 '[CH3:5][C:1](=[O:2])[OH:3].[CH3:6][OH:4]'
12 '>>'
13 '[CH3:5][C:1](=[O:2])[O:4][CH3:6].[OH2:3]'
14)
15
16# Build ITS graphs
17its_1 = rsmi_to_its(rsmi_1)
18its_2 = rsmi_to_its(rsmi_2)
19
20# Initialize the matcher, comparing element, charge, and bond order
21gm = GraphMatcherEngine(
22 backend='nx',
23 node_attrs=['element', 'charge'],
24 edge_attrs=['order']
25)
26
27# Test isomorphism
28are_isomorphic = gm.isomorphic(its_1, its_2)
29print(are_isomorphic) # True — they differ only by map labels
Example: Subgraph Search
Locate a smaller “reaction-center” ITS graph as a subgraph within a larger ITS graph:
1from synkit.IO import rsmi_to_its
2from synkit.Graph.Matcher.subgraph_matcher import SubgraphMatch
3
4# Core ITS graph of the first reaction
5core_its = rsmi_to_its(
6 '[CH3:1][C:2](=[O:3])[OH:4]>>[CH3:1][C:2](=[O:3])[O:6][CH3:5]',
7 core=True
8)
9
10# Full ITS graph of a second reaction
11full_its = rsmi_to_its(
12 '[CH3:5][C:1](=[O:2])[OH:3]>>[CH3:5][C:1](=[O:2])[O:4][CH3:6]'
13)
14
15# Initialize subgraph search engine
16sub_search = SubgraphMatch(
17
18)
19
20# Check if core_its is contained within full_its
21found = sub_search.subgraph_isomorphism(core_its, full_its)
22print(found) # True — the reaction center is present as a subgraph
ITS
The synkit.Graph.ITS package provides tools for constructing and decomposing Internal Transition State (ITS) graphs:
ITS construction
ITSConstructor— build an ITS graph from reactant/product NetworkX graphsReaction-center extraction
get_rc()— extract the minimal reaction-center subgraph from an ITSGraph decomposition
its_decompose()— split an ITS graph back into reactant and product graphs
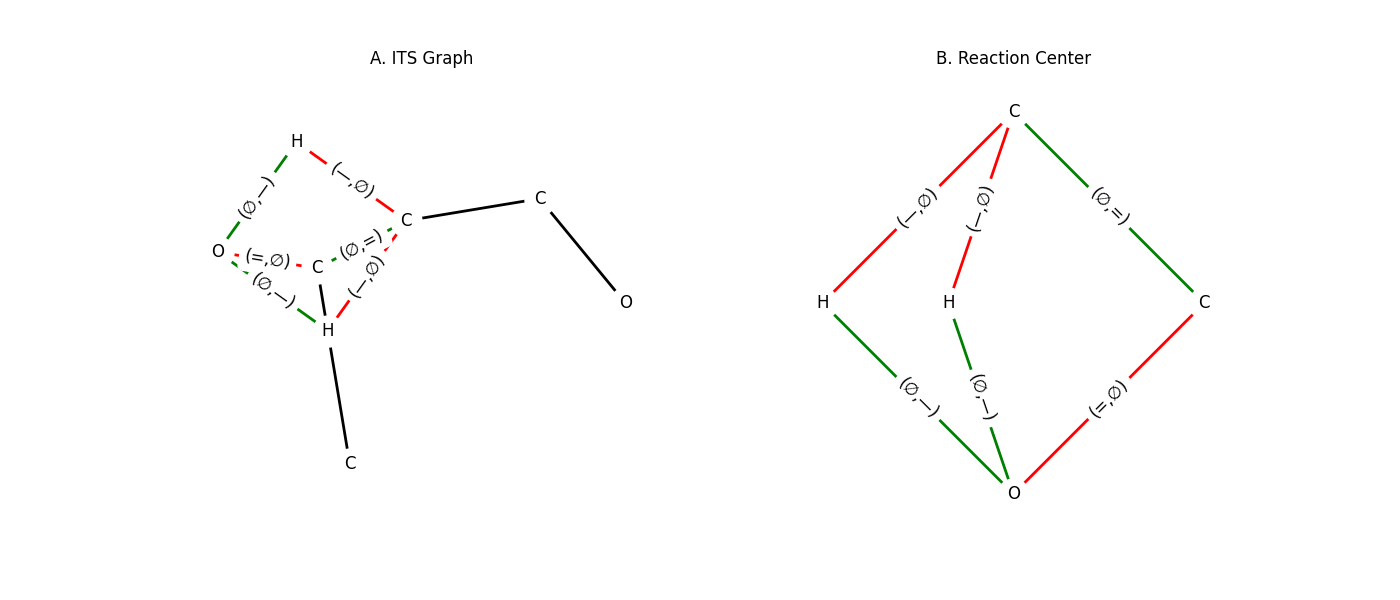
Example: Construct and Visualize an ITS
1from synkit.IO.chem_converter import rsmi_to_graph
2from synkit.Graph.ITS.its_construction import ITSConstruction
3from synkit.Graph.ITS.its_decompose import get_rc, its_decompose
4from synkit.Vis import GraphVisualizer
5import matplotlib.pyplot as plt
6
7# Parse the reaction SMILES into reactant and product graphs
8rsmi = (
9 '[CH3:1][CH:2]=[O:3].'
10 '[CH:4]([H:7])([H:8])[CH:5]=[O:6]'
11 '>>'
12 '[CH3:1][CH:2]=[CH:4][CH:5]=[O:6].'
13 '[O:3]([H:7])([H:8])'
14)
15react_graph, prod_graph = rsmi_to_graph(rsmi)
16
17# Build the full ITS graph
18its_graph = ITSConstruction().ITSGraph(react_graph, prod_graph)
19
20# Extract the reaction-center subgraph
21reaction_center = get_rc(its_graph)
22
23# Visualize both side by side
24vis = GraphVisualizer()
25fig, axes = plt.subplots(1, 2, figsize=(14, 6))
26vis.plot_its(its_graph, axes[0], use_edge_color=True, title='A. Full ITS Graph')
27vis.plot_its(reaction_center, axes[1], use_edge_color=True, title='B. Reaction Center')
28plt.show()
MTG Submodule
The synkit.Graph.MTG package provides tools for constructing and analyzing Mechanistic Transition Graphs (MTGs) from ITS reaction-center graphs:
MCSMatcherCompute maximum common substructure (MCS) mappings between two reaction-center ITS graphsMTGBuild a step-by-step MTG from a pair of ITS graphs and an MCS mapping
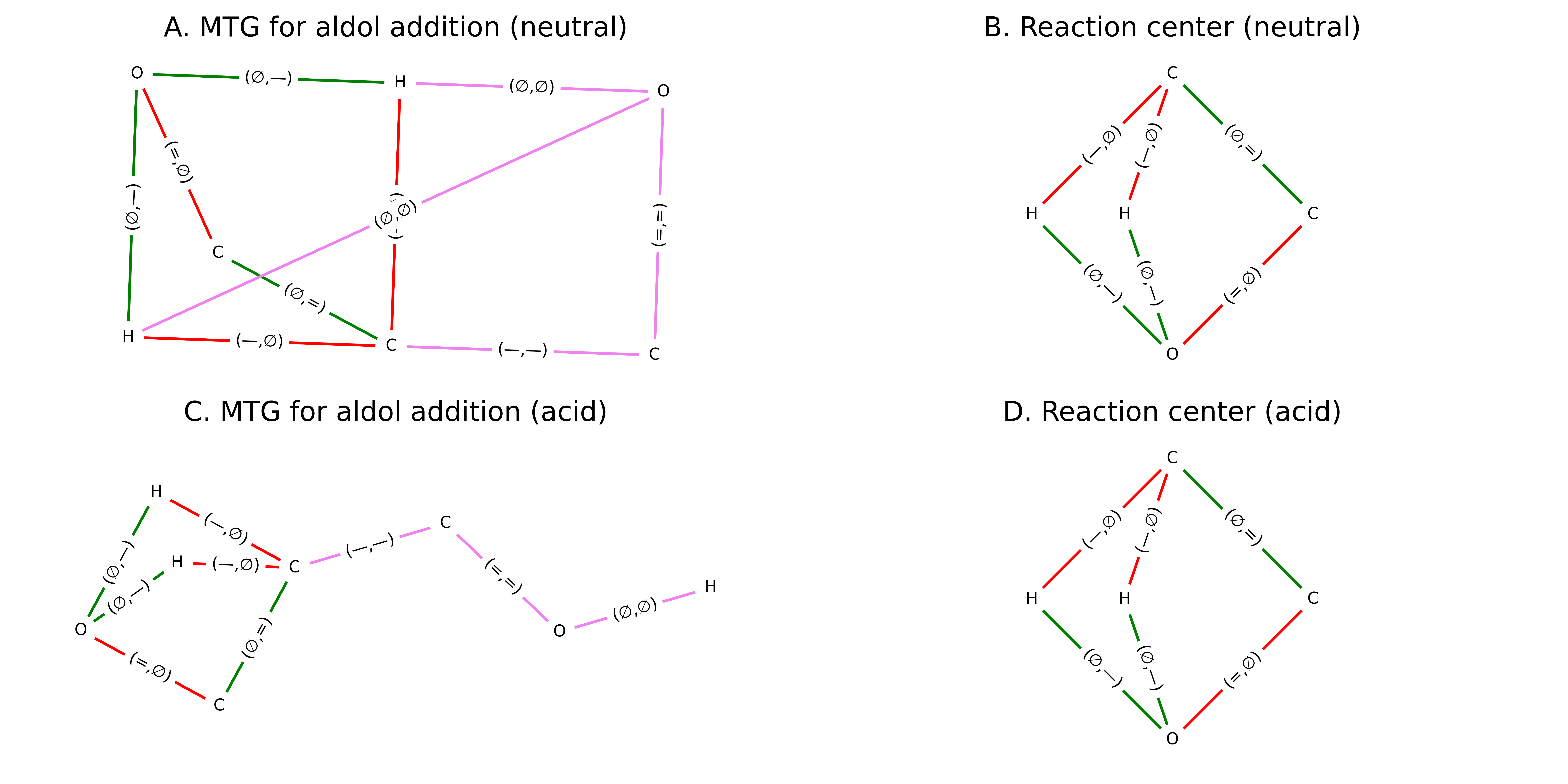
Example: Generate an MTG (with Composite Reaction Visualization)
This example builds two reaction-center ITS graphs, computes their MCS mapping, constructs the MTG, and then visualizes:
Each individual reaction center
The composite ITS for the overall mechanism
The final MTG
1from synkit.Graph.MTG.mtg import MTG
2from synkit.Graph.ITS.its_decompose import get_rc
3from synkit.examples import list_examples, load_example
4import matplotlib.pyplot as plt
5from synkit.Vis.graph_visualizer import GraphVisualizer
6
7
8data = load_example("aldol")
9
10mech_neutral = data[0]['mechanisms'][1]['steps']
11smart_neutral = [i['smart_string'] for i in mech_neutral]
12
13mech_acid = data[0]['mechanisms'][2]['steps']
14smart_acid = [i['smart_string'] for i in mech_acid]
15
16# neutral
17mtg = MTG(smart_neutral, mcs_mol=True)
18mtg_its_neutral = mtg.get_compose_its()
19mtg_rc_neutral = get_rc(mtg_its_neutral, keep_mtg=True)
20rc_neutral = get_rc(mtg_its_neutral, keep_mtg=False)
21
22# acid
23mtg = MTG(smart_acid, mcs_mol=True)
24mtg_its_acid = mtg.get_compose_its()
25mtg_rc_acid = get_rc(mtg_its_acid, keep_mtg=True)
26rc_acid = get_rc(mtg_its_acid, keep_mtg=False)
27
28# Visualize
29fig, ax = plt.subplots(2, 2, figsize=(16, 8))
30vis = GraphVisualizer()
31
32vis.plot_its(
33 mtg_rc_neutral,
34 ax=ax[0, 0],
35 use_edge_color=True,
36 og=True,
37 title='A. MTG for aldol addition (neutral)',
38 title_font_size=20,
39 title_font_weight='medium',
40 title_font_style='normal'
41)
42vis.plot_its(
43 rc_neutral,
44 ax=ax[0, 1],
45 use_edge_color=True,
46 og=True,
47 title='B. Reaction center (neutral)',
48 title_font_size=20,
49 title_font_weight='medium',
50 title_font_style='normal'
51)
52vis.plot_its(
53 mtg_rc_acid,
54 ax=ax[1, 0],
55 use_edge_color=True,
56 og=True,
57 title='C. MTG for aldol addition (acid)',
58 title_font_size=20,
59 title_font_weight='medium',
60 title_font_style='normal'
61)
62vis.plot_its(
63 rc_acid,
64 ax=ax[1, 1],
65 use_edge_color=True,
66 og=True,
67 title='D. Reaction center (acid)',
68 title_font_size=20,
69 title_font_weight='medium',
70 title_font_style='normal'
71)
72
73plt.tight_layout()
74plt.show()
Context graph
The synkit.Graph.Context submodule provides tools for expanding reaction center graphs to include nearest neighbors, enabling context‑aware analysis of reaction networks.
1from synkit.IO import rsmi_to_its
2from synkit.Graph.Context.radius_expand import RadiusExpand
3from synkit.Vis.graph_visualizer import GraphVisualizer
4
5smart = (
6 '[CH3:1][O:2][C:3](=[O:4])[CH:5]([CH2:6][CH2:7][CH2:8][CH2:9]'
7 '[NH:10][C:11](=[O:12])[O:13][CH2:14][c:15]1[cH:16][cH:17]'
8 '[cH:18][cH:19][cH:20]1)[NH:21][C:22](=[O:23])[NH:24][c:25]1'
9 '[cH:26][c:27]([O:28][CH3:29])[cH:30][c:31]([C:32]([CH3:33])'
10 '([CH3:34])[CH3:35])[c:36]1[OH:37].[OH:38][H:39]>>'
11 '[C:11](=[O:12])([O:13][CH2:14][c:15]1[cH:16][cH:17][cH:18]'
12 '[cH:19][cH:20]1)[OH:38].[CH3:1][O:2][C:3](=[O:4])[CH:5]'
13 '([CH2:6][CH2:7][CH2:8][CH2:9][NH:10][H:39])[NH:21][C:22]'
14 '(=[O:23])[NH:24][c:25]1[cH:26][c:27]([O:28][CH3:29])[cH:30]'
15 '[c:31]([C:32]([CH3:33])([CH3:34])[CH3:35])[c:36]1[OH:37]'
16)
17its = rsmi_to_its(smart)
18rc = rsmi_to_its(smart, core=True)
19exp = RadiusExpand()
20k1 = exp.extract_k(its, n_knn=1)
21
22gv = GraphVisualizer()
23gv.visualize_its_grid([rc, k1])
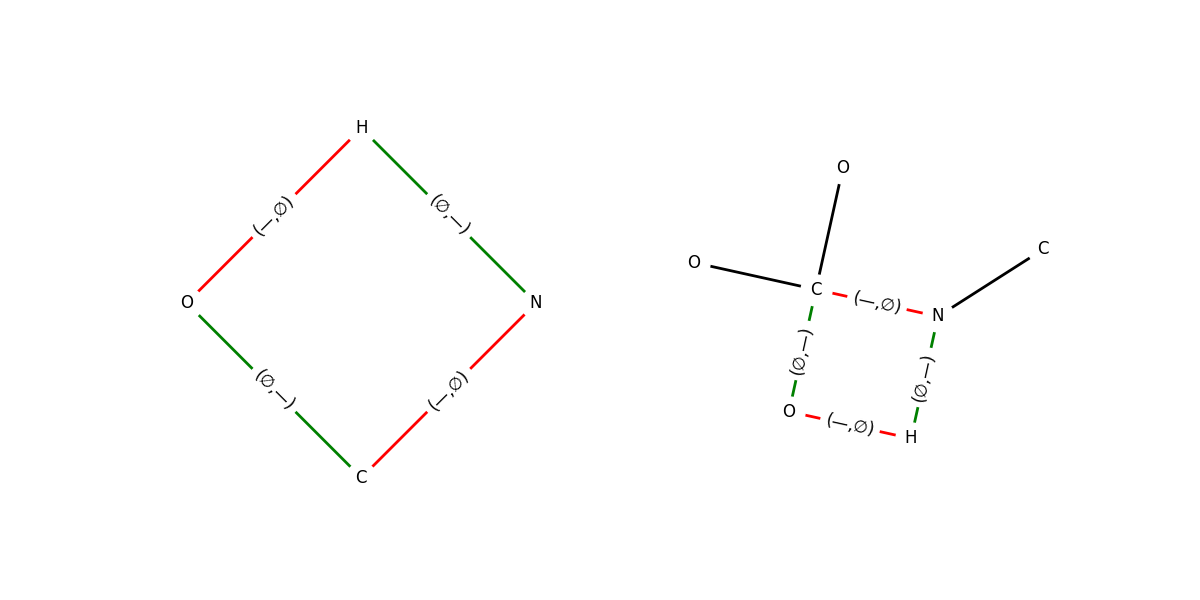
Figure: (A) Minimal reaction center subgraph obtained by contracting all atoms that participate directly in bond‑order changes. Nodes are colour‑coded by element; edges in red indicate bonds being broken, while edges in blue mark bonds being formed. (B) First shell ($k=1$) context expansion: every reaction center atom is augmented with all of its immediate neighbours.
See Also
synkit.IO— format conversion utilitiessynkit.Synthesis— reaction prediction & network exploration